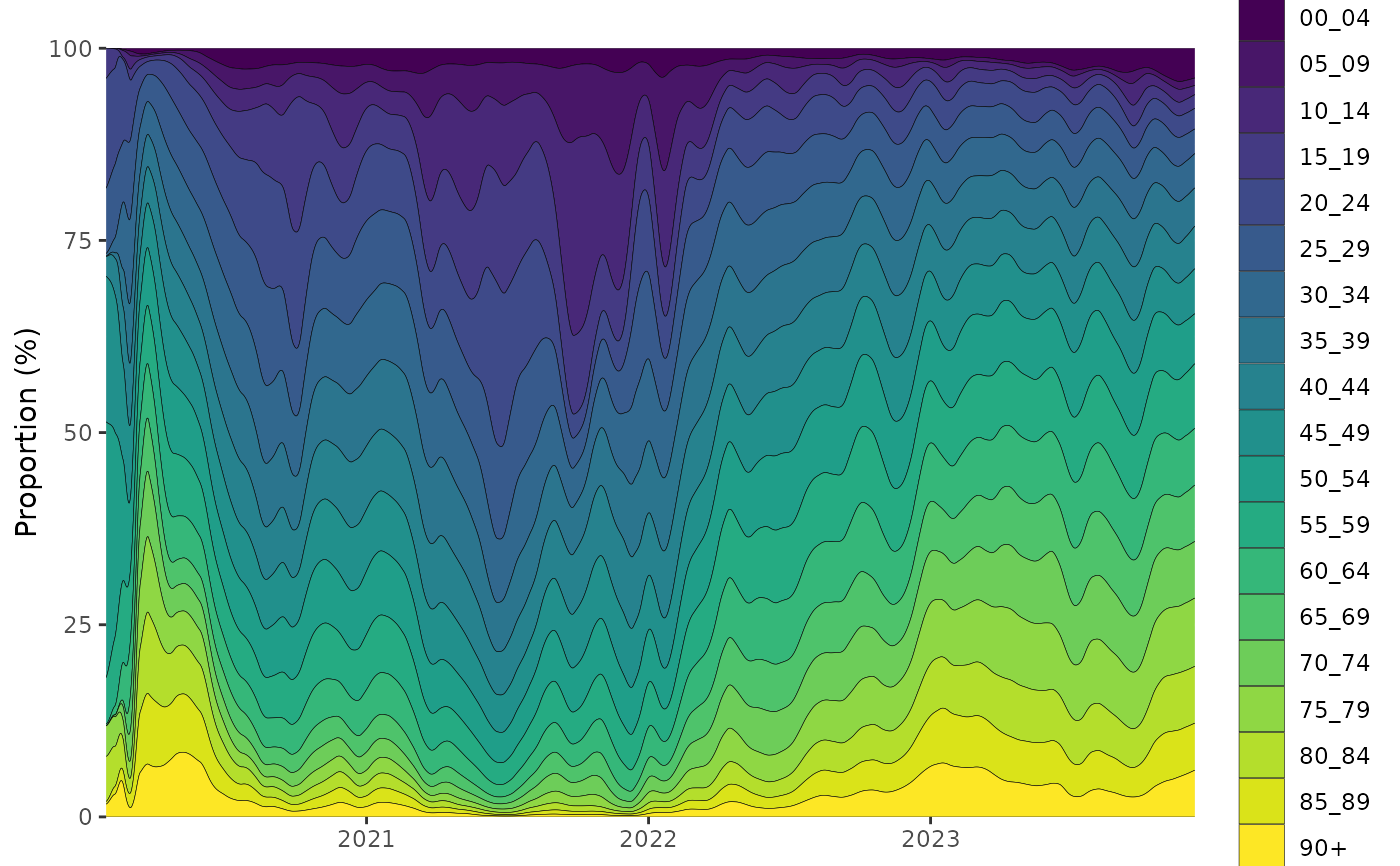
Plot a multinomial proportions mode
Usage
plot_multinomial(
modelled = i_multinomial_proportion_model,
...,
mapping = ggplot2::aes(fill = class),
events = i_events,
normalise = FALSE
)Arguments
- modelled
the multinomial count data
A dataframe containing the following columns:
time (as.time_period + group_unique) - A (usually complete) set of singular observations per unit time as a `time_period`
class (factor) - A factor specifying the type of observation. This will be things like variant, or serotype, for a multinomial model. Any missing data points are ignored.
proportion.0.5 (proportion) - median estimate of proportion (true scale)
Must be grouped by: class (exactly).
No default value.
- ...
Arguments passed on to
geom_events- mapping
a
ggplot2::aesmapping. Most importantly setting thecolourto something if there are multiple incidence timeseries in the plot- events
Significant events or time spans
A dataframe containing the following columns:
label (character) - the event label
start (date) - the start date, or the date of the event
end (date) - the end date or NA if a single event
No mandatory groupings.
A default value is defined.
- normalise
make sure the probabilities add up to one - this can be a bad idea if you know you may have missing values.
Examples
tmp = growthrates::england_covid %>%
growthrates::proportion_locfit_model(window=21) %>%
dplyr::glimpse()
#> Rows: 26,790
#> Columns: 12
#> Groups: class [19]
#> $ class <fct> 00_04, 00_04, 00_04, 00_04, 00_04, 00_04, 00_04…
#> $ time <time_prd> 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, …
#> $ proportion.fit <dbl> -13.433629, -13.178345, -12.898497, -12.600007,…
#> $ proportion.se.fit <dbl> 51.598289, 49.954079, 48.024633, 45.878749, 43.…
#> $ proportion.0.025 <dbl> 1.759164e-50, 5.698079e-49, 3.308357e-47, 2.991…
#> $ proportion.0.5 <dbl> 1.465037e-06, 1.891110e-06, 2.501801e-06, 3.371…
#> $ proportion.0.975 <dbl> 1.0000000, 1.0000000, 1.0000000, 1.0000000, 1.0…
#> $ relative.growth.fit <dbl> 0.24102860, 0.24048966, 0.23901181, 0.23680352,…
#> $ relative.growth.se.fit <dbl> 1.2309119, 1.2257057, 1.2114298, 1.1900979, 1.1…
#> $ relative.growth.0.025 <dbl> -2.1715143, -2.1618494, -2.1353470, -2.0957455,…
#> $ relative.growth.0.5 <dbl> 0.24102860, 0.24048966, 0.23901181, 0.23680352,…
#> $ relative.growth.0.975 <dbl> 2.6535715, 2.6428288, 2.6133706, 2.5693525, 2.5…
plot_multinomial(tmp, normalise=TRUE)+
ggplot2::scale_fill_viridis_d()